| designation: | D2-002 |
|---|---|
| author: | andrew white |
| status: | complete |
| prepared date: | August 15, 2025 |
| updated date: | August 17, 2025 |
abstract: There are over 10M research papers published per year and science has 227M total papers. FutureHouse and other companies are building AI scientists that are going to dramatically increase that count and become major readers and writers of science. The current publication system cannot sustain this and a new one must be built. I've written some of my thoughts on the topic below.
publishing for machines
Motivated by this essay from Seemay Chou and a discussion at SciFoo, I've been trying to put together my thoughts on "publishing for machines." As scientific agents become a major consumer of scientific literature, what changes should be made for writing and publishing articles?
reflections on publications
The idea of a scientific paper with citations was an incredible and prescient invention. It sounds obvious now, but the idea that written documents could be placed into a larger "corpus of science" via references is on the same level of the invention of writing. Writing allows us to store our thoughts and communicate them, but bounded by the length of the document. Attaching references in documents enables us to store our dialogues and observations across generations of humans, and now billions of pages of scientific text.
Scientific articles compose an unthinkably large non-linear document that is our distillation of scientific discourse over the last few hundred years. There are about 225,000,0001 of these documents and they are interconnected via about 3 billion citations. The only analogy is the internet, which has more documents and hyperlinks, but probably does not reflect our understanding of the universe as faithfully as scientific literature. Alongside the corpus of science, we have evolved natural language to better reflect our reasoning about science and the natural phenomena.[^2]
Some people have viewed the internet as a finite training resource, akin to fossil fuels2. In that metaphor, the internet is bunker oil--a thick gelatinous black goo which must be heated to even move through pipes. It burns dirty and is only fit for container ships and cheap cruises in international waters. Scientific literature is gas: distilled and efficient.
Except, scientific articles haven't had such an impact on training machines. There were a few mistakes made by our scientific ancestors. Papers are structured poorly and locked in PDFs, making them hard to parse, and scientists have broadly given up the rights to the articles, undermining the ability to actually navigate the citations that make up the scientific corpus. These mistakes have made the internet overtake the scientific literature for where timely and modern science is discussed. Issac Newton used the fastest and most efficient method to announce discoveries. So if he were alive today, he would be posting on arXiv or Twitter.3
Notice I do not complain about prestige journals or peer review. I believe those are separate issues from scientific publishing. They are professional and conscious choices made each day by scientists when they look at the journal titles in curriculum vitae and google scholar h-indices. If we replaced publishing overnight with a personal email lists, the subscriber count would be the prestige measure. Or, god forbid, we just go back to institutions, and your scientific career is set when undergrad applications are submitted at age 18.
So it would be wise to also address the structural issues of publishing as we undergo a transition towards having the producers and consumers become machines. And that motivates some of my thinking below.
machine readers
There are two sides to publishing for machines: the authors and the machine readers. Let's start from the reader-as-a-machine perspective. I make scientific agents that do keyword searches and download papers, just like a normal researcher would do. Just like normal researchers, these agents hit paywalls and have trouble downloading articles. They also have to deal with the fragmentation of a paper: the metadata is not available with the paper;4 the supporting information is a random set of links; and the articles come back as PDFs with figures, tables, text separated.
These are technology problems, easily solved: Make papers easier to access for machines at a URL. Provide the text/figures in a standard format. Provide the metadata at the paper, along with a single zipped archive of supporting information.
A larger issue for machines as readers is the actual the content of papers. Papers follow a certain formulaic narrative regardless of the actual scientific process that lead to them. Only successful results are reported. The order of the manuscript is rearranged to be as compelling as possible. Papers often contain multiple claims and hypotheses, being more a grouping of experiments/theory into one topic. Papers almost always conclude in an optimistic definite result. The actual skepticism of science is hidden behind peer-review and conferences. There are odd appendages, like supporting figures from peer review or interesting side-notes included. Papers are almost never retracted or refuted, except in cases of extreme negligence or outright fraud.
Papers just do not reflect the scientific process and they are written to conform to a professional standard that serves little purpose anymore.
The easy answer to making papers for machines is to remove human narrative: just make the papers the data, the code, and an accepted/rejected hypothesis.
I disagree with simple approach. Humans provide an extremely valuable role in distilling and discussing. I don't want to erase humans from the narrative. It's also tempting to just separate the data from the narrative, which I also find distressing. There is no such thing as "data" without explicit assumptions and choices that are based on some kind of research direction.
Ok - with the caveats and setting, here's my moderately thought out vision.
future papers
I want these attributes of papers for machines: (1) low latency from results to paper; (2) details on the purpose (hypothesis or goal) of the paper; (3) "raw-ish" data with an explicit description of methods and the minimal processing done; (4) a list of results, along with confidence, concordance with existing literature, and analysis (natural language is fine) that supports them; (5) discussion with actual commentary about how the paper changes the author's perspective on the domain. I'll define them in more detail below.
The pieces do not all need to be present. A paper without results is a data generation paper. A paper with a discussion, but nothing else, could be a peer review. Results without data is a reanalysis of previous papers. A hypothesis only is an opinion piece.
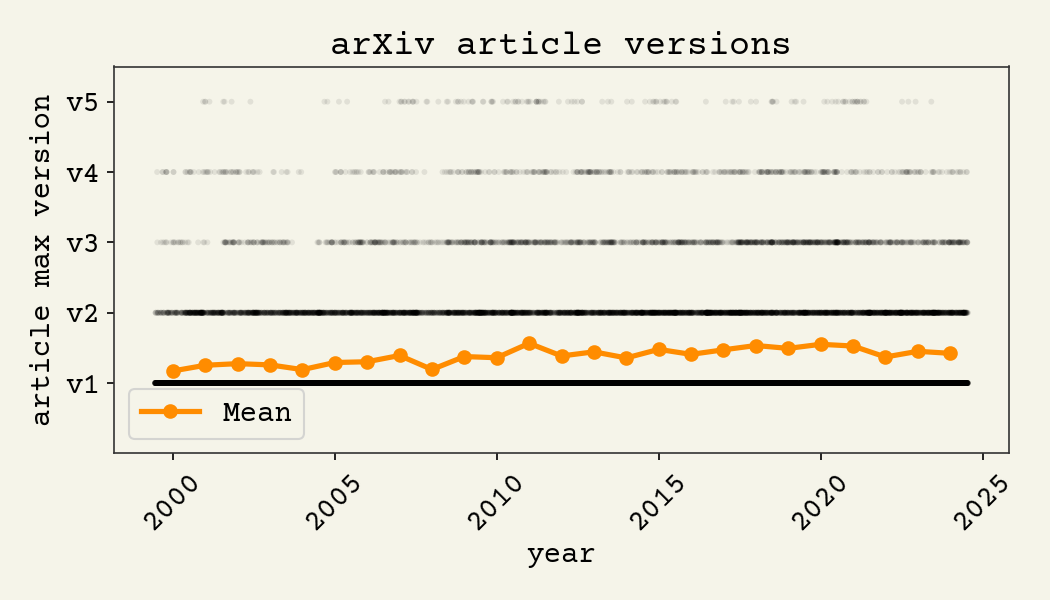
latency is a big change. Research publications started as letters to aggregating journals because it was the fastest mechanism for one-to-many communication. We need to recapture that speed. Increasing the speed of publishing will lead to people making mistakes or updating datasets or analysis. That's ok. Just upload a new version. Scientists already utilize the versioning feature and upload new versions of preprints with minimal fanfare, as shown below. The figure shows how many updates are made to preprints from a sample of 50,000 arXiv preprints.
The normal scientific literature only supports retractions or corrections, which are considered embarrassing. About 0.02% of scientific literature has been retracted.5 The ability to seamlessly and quickly update preprints on arXiv has revealed that scientists would be correcting papers at a closer to 50% rate.
hypothesis: a sort of guiding research purpose must be stated at the beginning of paper. Likely a hypothesis. This is the most human activity in science: the choice of what questions are important. An explicit research purpose provides essential context for decisions made in the paper. Assume I have a hypothesis that protein monomers are more common in the cytoplasm than the cell membrane. If I exclude proteins localized to the nucleus, it requires no explanation in my analysis. It's unrelated to the cytoplasm and membrane. If my hypothesis is instead that intrinsically disordered proteins have less glycosylation, it would be unobvious why proteins localized to the nucleus should be excluded. It would require some explanation. A hypothesis at the top provides that extra context for subsequent decisions in data, processing, and analysis.
data: raw data is overrated in my opinion. The first stage of converting raw data to processed data truly requires the person that collected the data. A person disconnected from the experiments cannot make the processing decisions. Maybe I know my signal on the NMR from experiment 2 went bad because my solute crashed out, so I should just cut the data at 6 hours. Maybe it's data from the LHC and you literally cannot publish the raw data because it's so large. The person, or agent, should decide when the clear-cut steps are done and then mark that data as "raw-ish" and process no further.
results: the results should be claims supported by the evidence and be couched in existing literature. This is the actual increment of science. This is the chance to provide negative or contradictory results as well. The description of the analysis is also essential, and it might be code or text. The beautiful thing is that code and language are near inter-convertible now, with the advances in code-writing large language models.
discussion is the update to the world model of the author. What did they believe would be the result based on known literature? Which results did not fit that belief? In the counterfactual before doing the experiments, what minimal information could have been in the world model of the author to predict these results from their understanding of the system being studied. Maybe it's no new information. That's ok. The discussion should be opinion, because it is clearly the viewpoint of the authors.
peer review
Science is an adversarial process. Each venue accepting submissions has to deal with an enormous amount of spam.6 There is a strong demand to get easy content into the scientific domain.7 Behind the scenes at arXiv, they are continuously fighting against AI-generated slop papers. It is a necessary process that is generally invisible to scientists. Automated peer review should address this onslaught and serve as the front-line for publishing research.
Human peer review of literature serves an extremely important purpose: providing opportunity for private debate with consequences. Much of the true debate of science occurs in the peer review. It is high-stakes, so that participants must state their opinions. That process is valuable and should not be hidden in peer review. Automated peer review should check for best practices, correct citations of the literature, and adherence to the format and rigor of science. It should be decoupled from the debate of science. That debate should exist in some other format though, not private correspondence. Peer review to debate science could live in separate papers, but I don't have strong conviction on this or know how to solve for peer review.
what we don't need
The last year has led me to re-evaluate "exactly reproducible papers." Previously, I supported the idea of papers that could be one-click repeated via technology like containerization, continuous integration, version control of analysis code, and sharing of data. I have done this in my own papers. I found it to be a large amount of effort and brittle, requiring a lot of finesse.
I no longer believe this level of work is required. Assuming the analysis is standard enough, modern scientific agents know how to load data, parse it, do pretty complex calculations, and generate compelling figures. Of course, it is nice to have all of these things automated in one clean repo with a README. Yet I don't think it needs to be an essential part of future papers.
Negative results are constantly brought up in discussions like this. I'm actually not strongly in favor of publishing negative results. There are many reasons why a result could be negative. It could be technical failures. It could be not enough troubleshooting was applied. It could be that we didn't get good traction after 2 weeks and decided to just move on to a more promising approach. It could be so many things! Yet, a positive result shows that all parts of the process mostly worked. Actually sufficiently justifying a negative result by checking all parts of the scientific process carefully is more work than a positive result.
My point is that "just publish negative results" is actually a pretty nuanced and complex task, and so I don't think it should be enforced as a policy in scientific publishing.
One approach to negative results that gets at this fundamental limitation of negative results is being tried by Arcadia. Their "Icebox" project reports negative results, but they are marked with a reason. Examples are "strategic misalignment" or "lack of infrastructure." It's the best approach to publishing negative results I've seen, since it shows why it might be negative.
journals
There will need to be some curation for human readers of this system. There could be "virtual journals," that simply curate content from the fast moving machine publications. These could be operated by AI, having systems that surface papers without human intervention. It may be tempting to have these things be personalized, but I think it's still useful to have a shared curation for people to discuss papers.
Who will run the machine publishing platform? I don't know. I know some readers of this article will jump to technology like a blockchain and decentralized science. I think a decentralized system could solve some technology challenges, but some kind of decision making body that decides how automated review is done and stewarding resources is necessary. Wikipedia could be one success story to look up to. Another is arXiv and openrXiv. More radical ideas on publishing are also worth looking at - like research hub and Arcadia.
I also believe professional societies and commercial publishers should participate in this process. There are real costs and value in curating scientific literature. Just, I want the commercial model to look more like Google or Twitter, where their value is in the curation of content, rather than charging for access of the underlying data.
authors
What do authors get out of this system? Less friction to publish is one. Low latency means hours or minutes to publish, not months. Another benefit is that novelty and impact are not accounted for in such a system, except post-hoc. In some ways it's more work. Just publishing in a high impact journal is not enough to guarantee readership.
There will be citations from this machine publishing platform. If they are viewed as "first-class" citations by google scholar, then it would eclipse traditional citations rapidly. Since that is the silently agreed upon measure of merit in most academic institutions (especially outside the US), this system would be viewed favorably even by non-participants. There will be huge citation inflation.
It will become much harder to wade through the literature. Already, I rely on my own research agents8 more than traditional scholar search products. Machine publications should easily outstrip human publications and hit 10M or 100M per year of articles quickly.
conclusions
A system for machine-generated scientific publications must be built soon. Already, at FutureHouse we have something siloed like this that is starting to generate a parallel corpus of scientific knowledge. I'm sure other members of the AI scientist race are doing the same. If no public facing option can be built, it will be strongly detrimental for the scientific community.
reflections
These ideas are generally not new. There have been many attempts to increase speed of publishing, make open access default, and remove any novelty or impact requirement. Examples include journals like Scientific Reports, ACS Omega, and Frontiers journals. They have lower-quality uninteresting papers, generally, and I have an internalized negative signal for papers from those journals. I can't remember citing something from one of these journals, but our literature research agent (PaperQA2)8 loves to read these journals--which I'm still confused about. Maybe I'm just old and stubborn.
If you randomly select an arXiv or biorXiv preprint, it also is likely of below-average quality relative to a peer-reviewed paper. There are some weird and low-quality papers there. However, 99% of influential AI research is on arxiv now and it's been a huge success for the field.
I'm not sure what to learn from these observations on past experiments. Maybe execution matters? Maybe community buy-in matters? Maybe we need to decouple curation from publishing and my observations are from the unnecessary coupling.
Footnotes
-
I don't have a great reference, but the search bar on semantic scholar updates every day with this number ↩
-
Famously said by Ilya Sutskever at a talk at NeurIPS 2024. ↩
-
Of course, I mean "Twitter" in the platonic ideal form of social media, not literally the social media site formerly known as Twitter. Credit to Chris Wilmer for explaining this to me. ↩
-
you can go to crossref or semantic scholar for this ↩
-
For example, Zenodo had to lock down to avoid this: https://support.zenodo.org/help/en-gb/2-safelisting-spam/141-what-content-do-you-consider-spam ↩
-
Like getting homeopathy articles into journals: https://www.nature.com/articles/s41598-024-51319-w ↩
-
Skarlinski, Michael D., et al. "Language agents achieve superhuman synthesis of scientific knowledge." arXiv preprint arXiv:2409.13740 (2024). ↩ ↩2