| designation: | D1-011 |
|---|---|
| author: | andrew white |
| updated date: | November 26, 2025 |
abstract: The Edison API can be used to do "for each" style intelligence tasks. Here I show how to convert powerpoint presentations to scientific claims and then diligence each claim to see if they are contradicted in literature/patents/clinical trials.
using edison API
I've been building AI agents for a while now. They're most interesting when deployed at scale. Projects like Robin1, which makes drugs, and Kosmos, an "AI Scientist", only work because we can run thousands of AI agents at once.
We've had this for a while at FutureHouse. We first built out our literature agent, PaperQA, on an API. We ran many experiments using the API, like wikicrow. Wikicrow is a website with a wikipedia article for all 20k protein-coding genes in the human genome. We once ran a crazy combinatorial meta-analysis on every peptide on every behavior in every animal model. This led to a big matrix showing which peptides with missing experiments are predicted to have a specific behavior effect in humans. It was cool, but when we tried to submit it to a journal they asked if it was real or not. Then we realized it would be hard to know if any of these predictions are true.
This is the problem with literature experiments: a hypothesis cannot be published unless you provide experimental/computational evidence for it. So we actually didn't continue a lot of projects based on hypotheses/predictions at FutureHouse, since much our output was focused on papers. To prove any of what we built was useful, we ended up backing ourselves into building a world-class evals group (led by Jon Laurent). We built things like LabBench2, BixBench3, and even ether04 because we had to find a way to measure AI Agents for Science.
Now at Edison Scientific, we aren't trying to publish papers. The mission of Edison Scientific is to accelerate scientific discovery, directly. So we put our agents on an API and let people use them at very high rates. I'm excited to see what people build with it!
diligencing a powerpoint
I want to show a demo of what you can do with this API now. Let's say I was given a powerpoint presentation (or 100) and want to check every claim in the presentation to see if it's plausible. We could approach this by analyzing primary data using Finch, our data analysis agent, but I want to try an easier approach.
Let's just take every claim and ask PaperQA "Has anyone ever contradicted this claim before, and how compelling is that contradicting evidence?" This will give a quick test on if any claims seem outlandish and we can dig more into them. We built something just like this and showed humans usually agree with the analysis in our PaperQA2 paper.5
So I want to go through these steps:
- Convert a powerpoint presentation into a list of claims
- For each claim, check if it is contradicted in literature/patents/clinical trials
- List the most problematic claims
Here's my presentation I'll be using in the demo (it's two slides):
extracting claims
Let's start with step 1. First, make sure the powerpoint is in a PDF format. Then I just asked Claude to write code to do step 1 (PDFs to claims):
The code that came out was fine (I'll link to it all at the end) and it returned the resulting claims from my presentation:
GGA DFT can model water at room temperature and get density correct
FDA approvals of small molecule KRAS inhibitors are generally covalent binders that hit cysteine mutation
You can easily use OpenAI, Gemini, or any other LLM for this part. They all support directly uploading PDFs and they're quite capable of extracting claims from documents.
edison API
Now let's see how to use the edison API to check these claims. First, let's make a rubric we will use:
rubric = [
"Strong confirmatory evidence",
"Confirmatory evidence",
"No evidence one way or another",
"Some contradicting evidence",
"Strong condracting evidence"
]
rubric_text = "\n".join([f"{i}: {s}" for i,s in enumerate(rubric)])
This will make a nice rubric we can use in our prompt, like so:
Important: Start your answer response with an integer from 0 to 5 according to this rubric:
0: Strong confirmatory evidence
1: Confirmatory evidence
2: No evidence one way or another
3: Some contradicting evidence
4: Strong contradicting evidence
Now we can call the API
from edison_client import EdisonClient, JobNames
client = EdisonClient()
task_data = {
"name": JobNames.LITERATURE,
"query": query,
}
task_response = client.run_tasks_until_done(task_data)
print(task_response[0].answer[0])
# 4
We got back strong contradicting evidence, which is definitely correct. This is an insane claim about molecular dynamics. You can read the explanation/citations for the strong contradictory evidence here. Here's a quote from it:
and it cited 9 articles about the topic.
packaging it up
Now let's package it up into a nice script. I gave this to Claude (opus-4.5)
Then it will call the Edison API with at most 5 simultaneous calls. It should have a pleasant display of progress - like a carrot pointing at each claim. Then when a response is returned, show a nice icon based on the response value.
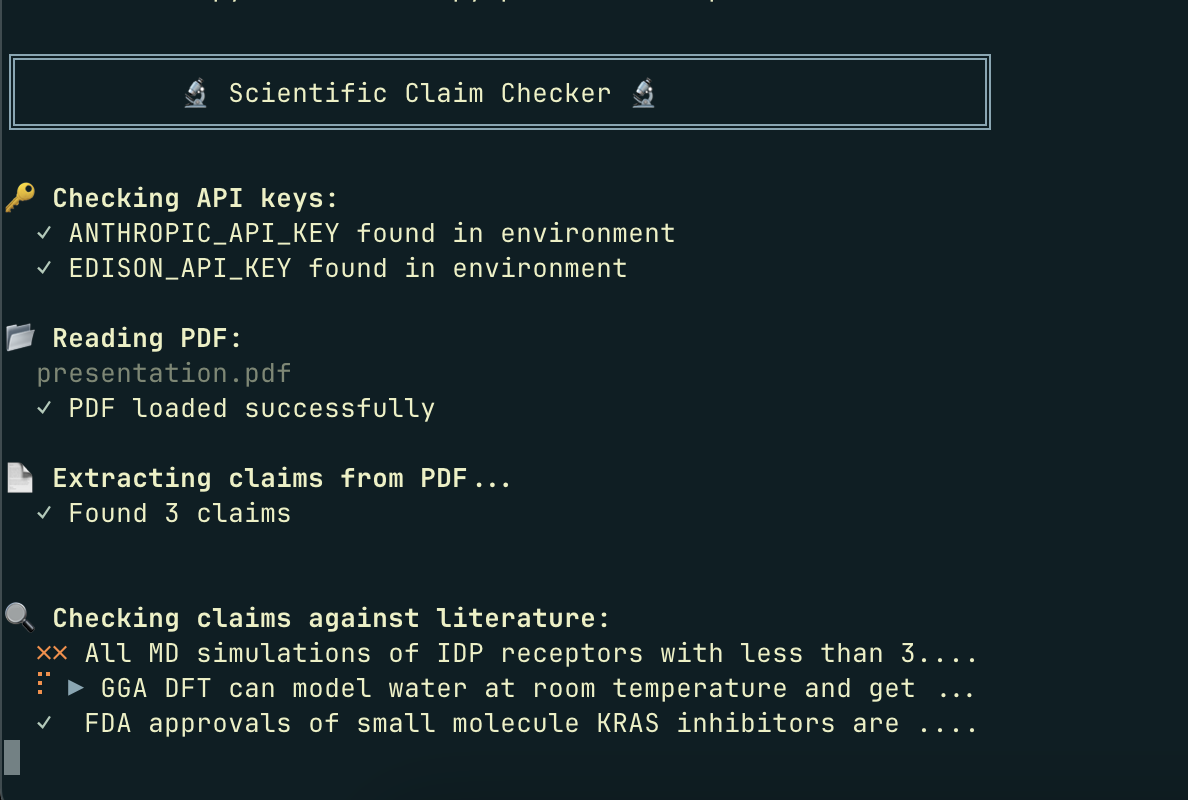
And it gave me a pleasant script to run. I executed it on my presentation above:
uv run python checker.py presentation.pdf
and you can see the output as it loaded
It is indeed true about the FDA claim. The GGA claim is definitely contradicted. These are relatively simple claims and could be treated by a simple Google search; they're just for illustrative purposes.
some install instructions
If you want to try this yourself:
- Get an API Key from Anthropic
- Get an API Key from Edison
- Go install uv
- Download the script
- Run
uv run python checker.pdf [PDF input file]
Hope you find it interesting!
discussion
I actually don't run into the urgent need to diligence hundreds of powerpoint presentations. I got this idea from discussing the process of vetting a company for acquisition with someone who does this for a living. I guess a ton of company scientific findings are locked away in powerpoints and the acquisition period for reviewing those documents is quite short. I'm not sure if this approach would be provide a lot of signal, but it's nice demonstration of the kind of tasks that you can undertake with scientific agents on API.