| designation: | D1-008 |
|---|---|
| author: | andrew white |
| status: | complete |
| prepared date: | February 2, 2025 |
| updated date: | February 19, 2025 |
abstract: Image duplication has been a powerful signal for detecting scientific fraud. Image duplication is irrelevant in many fields. Here I propose a few other ideas of signals for fraud and find one that significantly correlates with retracted papers. The idea is to validate citations, rather than papers, by considering both the citing paper and the cited paper simultaneously with large language models. Assuming no issues with accessing full-text papers, it would cost about $1 per paper to run this check and thus we could cover all arxiv submissions for $25,000 per month, all new peer-reviewed papers for $500,000 per month, all literature for $250M, and all of Wikipedia for $130,000. Although my signal is relatively weak, the scale and quality of large language models means that we may soon have a new suite of tools for detecting fraudulent papers.
finding new signals for scientific fraud
I think we've been quite lucky with scientific fraud. One of the most tempting ways of faking results is duplicating parts of images, especially in biology. Biology is one of the highest-stakes fields. Fraud can lead to wasted effort on developing drugs and running human trials. Luckily image duplication is visible from figures alone and is unambiguously fraud.1
The alternative methods of detecting scientific fraud are much more difficult. Sometimes people are egregiously sloppy with statistics.2 Or they make such insane claims that the field has to investigate.3 Or people observe the fraud directly and try to whistleblow it, but that is actually pretty difficult as a PhD student or junior researcher.
We got a clue recently of the state of fraud in fields that don't involve image duplication with a recent preprint. Richardson et al.4 looked at materials science papers to check if the microscope instruments reported in papers could have plausibly made the images reported. Like if a paper methods claimed to use a Sony microscope, but all the pictures said Lyko in the corner. They found this inconsistency in about 25% of papers they analyzed.5
25% may sound like a shockingly high number. It is much higher than what we see in image duplication,6 where estimates have put it round 6%. However - that recent analysis has come after the field has to grapple with widespread image duplication and now screening has become part of the editorial process. Prior to that, around 15% of papers had duplicated images.6 So maybe 15-25% of papers containing fraudulent images is expected? A depressing statistic.
The denominator is also hard in these discussions. Namely, what papers are you running considering in these analysis? It is hard to get full-text papers for automated analysis. The publishers that hold onto full-text papers the strongest are often the ones that sell access to their full-text papers because they are valuable. The publishers that rely on collecting publication fees are not so concerned with blocking people from accessing their full-text. So you see an inverse correlation with ease of access and tier of journal.7 Thus the 25% could be an over-estimate.
Ultimately though, fraud is a bit tangential to what we care about. Scientists care about if a reported research result is "true": can you reproduce it. Fraud implies no reproducibility but no fraud does not imply reproducible.
Reproducilibility is in an even worse state though, with dedicated efforts showing > 50% of papers are not reproducible.8
What about just using primary data and avoiding papers? That's an argument I hear a lot: go to the root of data, the primary data. Well it turns out that papers which are retracted and fraudulent also are depositing fake data into primary data sources. Bimler found that retracted papers that deposit corresponding crystallography data were depositing fake structures too.9 They did this via duplicating real data and then lightly editing it to fit their results. Thus, I expect across other fields people are depositing fake data into primary databases on gene expression, protein structures, etc. Some, like the Protein Data Bank, have very good filters for this and structures have been removed.
Ok - background out of the way. Can we find a new signal that spans domains and detects fraud faster, like image duplication?
automated methods to validate the literature
so the goal is is find a new signal to detect fraud automatically across all of science. Here are attributes of a "universal fraud signal:"
- it has a low false positive rate
- it can be computed on papers from any field
- it correlates with non-reproducible work or fraud
There is no perfect way to test such a signal. My test here will be if it looks reasonable and correlates with retrospectively predicting if a paper was retracted -- namely could it have predicted retractions that occurred in the past.
hypothesized signals
Here are some ideas that can fit this:
- plagiarism
- self-citations
- citations to irrelevant papers
- citations that are mischaracterizations of the cited work
- reproducing numerical analysis in the papers
From this list, plagiarism is already pretty well-explored and detected by publishers. Self-citations is interesting. It's not "fraud," but my point here is that it could be a signal that surfaces fraudulent papers.
Citation character is interesting. A claim - like the sentence "Short people live longer (Foo et al., 2000)" - could be citing an irrelevant paper by Foo. Or Foo could be a paper that disagrees with the claim. The citation to an irrelevant paper can arise in so-called "citation rings," where people will cite your articles for some amount of money to boost your publication metrics. Those should be detected with this method, since presumably they are randomly inserting citations to earn money from the citation ring. On the other hand, a mischaracterization could represent sloppy work. Maybe that is a signal.
Reproducing the numeric analysis is a final option. We're close with large language models, but there are a lot of problems of obtaining the primary data to repeat analyses and there is much more room for debate on what is necessary to be considered "reproducible."
Based on this discussion, here's what I tested today:
- self-citations
- citations to irrelevant papers
- citations that are mischaracterizations of the cited work
data
I downloaded the Retraction Watch database as hosted by CrossRef.10 It contains retracted papers as DOIs. I then found a matching article for each retracted paper. The match had to be published in the same journal and same year. And yes, I also checked to make sure the matched article wasn't itself retracted. From this set of paired retracted/non-retracted articles I downsampled to 341 pairs for which I could get full-text (from open source or preprints). These 682 articles were then analyzed according to the three signals above.
method
I wanted a method that does not depend on the article being in the scientific corpus, because I want to use this signal on grant proposals or technical reports. So the input is a PDF. Since parsing PDFs is fraught, I just put the entire PDF into Google's Gemini model11 and have it parse out cited claims and the reference being cited.
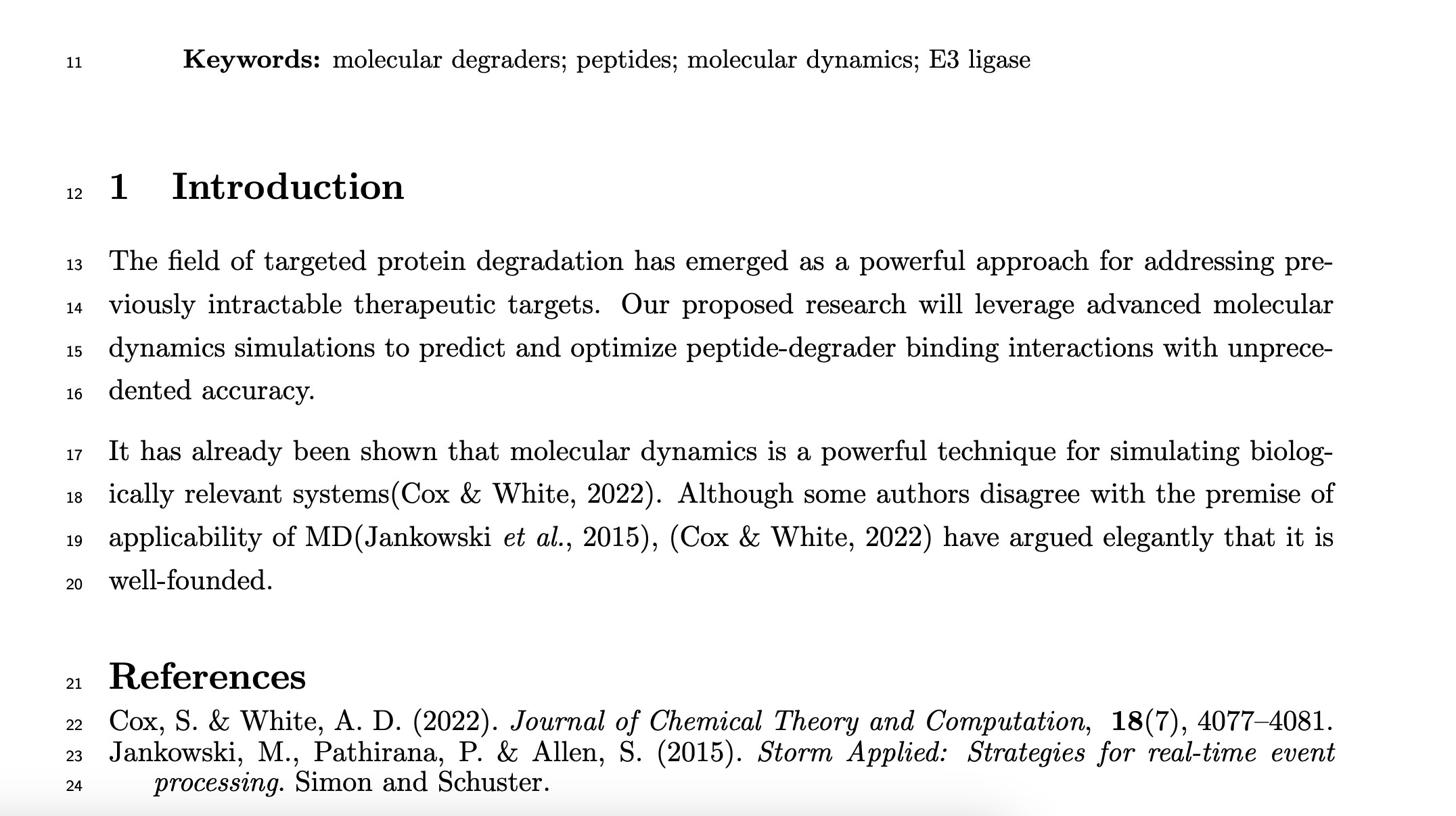
Here's an example image of my starting PDF:
And here is what is parsed from that PDF:
"Cox & White, 2022": {
"title": "Symmetric Molecular Dynamics",
"authors": [
{
"family": "Cox",
"given": "S."
},
{
"family": "White",
"given": "A. D."
}
],
"year": "2022",
"journal": "Journal of Chemical Theory and Computation",
"identifiers": null,
"url": null,
"key": "Cox & White, 2022",
"self": true,
"citation": "Symmetric Molecular Dynamics. Cox, S., White, A. D.. Journal of Chemical Theory and Computation.",
"contexts": [
{
"citation_key": "Cox & White, 2022",
"context": "It has already been shown that molecular dynamics is a
powerful technique for simulating biologically relevant systems(Cox & White, 2022).",
"category": "support"
},
{
"citation_key": "Cox & White, 2022",
"context": "Although some authors disagree with the premise
of applicability of MD(Jankowski et al., 2015),
(Cox & White, 2022) have argued elegantly that it is well-founded.",
"category": "contrast"
}
]
},...
The fact that I can get this information out with a single LLM call is amazing. Building systems that can analyze arbitrary PDFs and do this extraction has consumed probably almost 1,000 PhD dissertations. I was able to just do a single LLM call to get that data.
Now the hard part: how do I check the validity of these claims? I have to actually get the PDFs of these cited references. This is hard and complicated. I try preprint servers, downloading open access from publisher websites (which block automated downloads), and by-hand clicking on download links. It's hard and that's why I feel there is so little innovation on topics like this. Anyway, a challenge for another day.
Now once I have the PDF, I can use Gemini's ability to just eat PDFs from Google Cloud Storage buckets to get a nice clean prompt. So I ask
{PDF of Cox & White as GCS link}
Assess the agreement of the claim(s) below with the primary source above.
The primary source was cited in these claims:
"Although some authors disagree with the premise of applicability of MD(Jankowski et al., 2015), (Cox & White, 2022) have argued elegantly that it is well-founded."
"It has already been shown that molecular dynamics is a powerful technique for simulating biologically relevant systems(Cox & White, 2022)."
Classify the correctness of these claims in relation to the primary source according to the following options:
A) The cited statement is supported or consistent with the primary source.
B) The cited statement mischaracterizes or is contradicted by the primary source.
C) The cited statement clearly is irrelevant to the primary source.
Answer in the following format:
{
"reasoning": "reasoning about choice",
"choice": "single letter choice"
}
If there are multiple claims, please provide a single choice representative of all contexts.
And the output
{
"reasoning": "The paper focuses on a novel method for molecular dynamics simulations that incorporates symmetry constraints.
While the paper mentions the use of MD for biological assemblies and crystal structure prediction, it does not explicitly
claim that MD is a powerful technique for simulating *all* biologically relevant systems. The second claim is difficult to
assess without the context of Jankowski et al. (2015), but the paper does not appear to directly address the general
applicability of MD or argue against any specific criticisms of it.",
"choice": "B"
}
So we can conclude that the paper mischaracterized Cox & White, which is indeed correct. Cox & White was about ideal particle simulations with symmetric constraints. It is not quite irrelevant, because there is some mention of biologic relevance in there. The reasoning is also good.
results
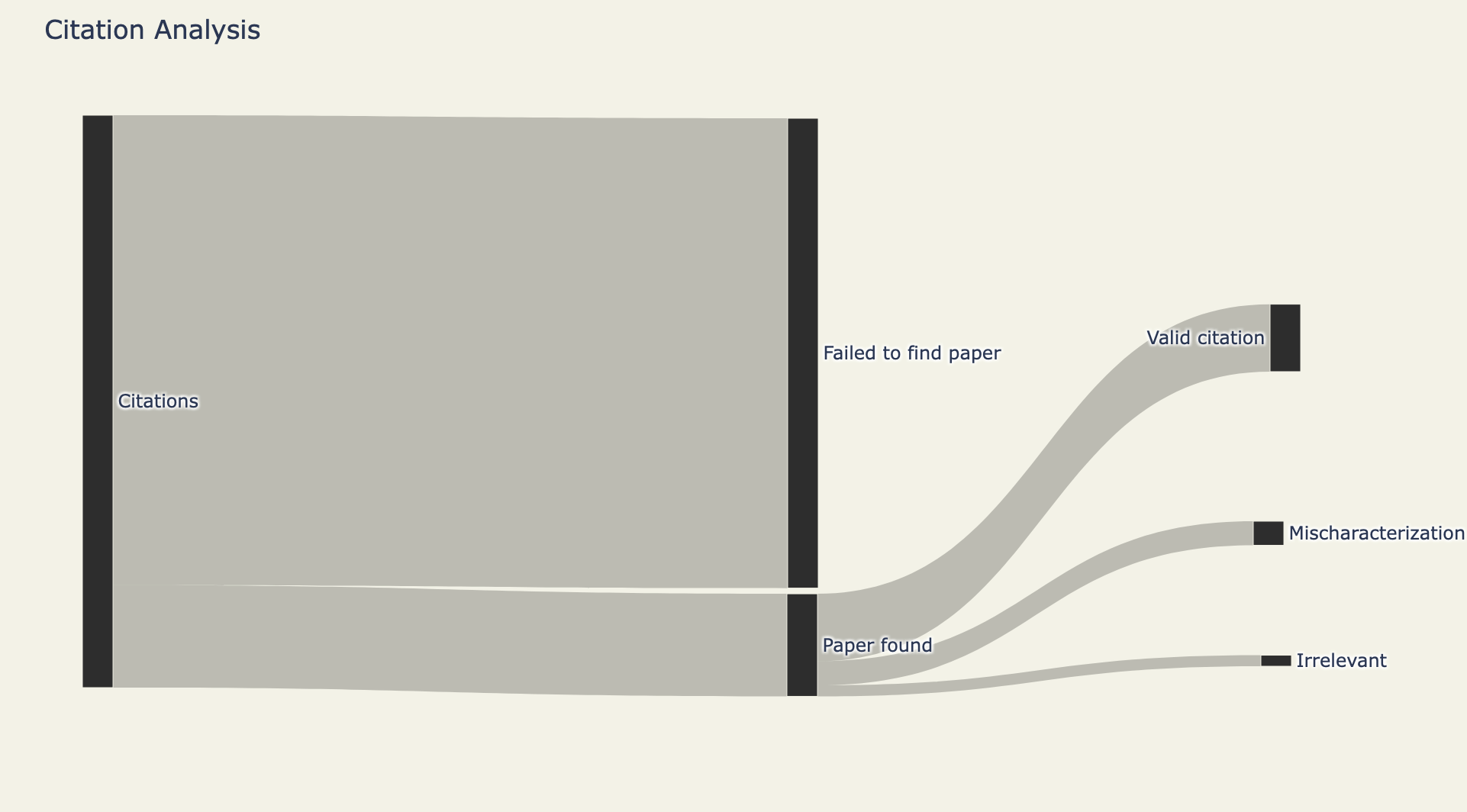
Now I ran this on the 682 papers mentioned above. Here's a Sankey diagram of how the analysis went:
I considered 20,600 references from the 682 papers. From those 20,600, I was able to get 3,700 full-text papers. Part of the reason is access to papers. Another is that I used a strict criteria to avoid leaking non-matched articles - because accidentally grabbing an incorrect article or supporting information on accident would increase the "irrelevant" article count. From those 3,700 citations, 2,430 were valid citations.
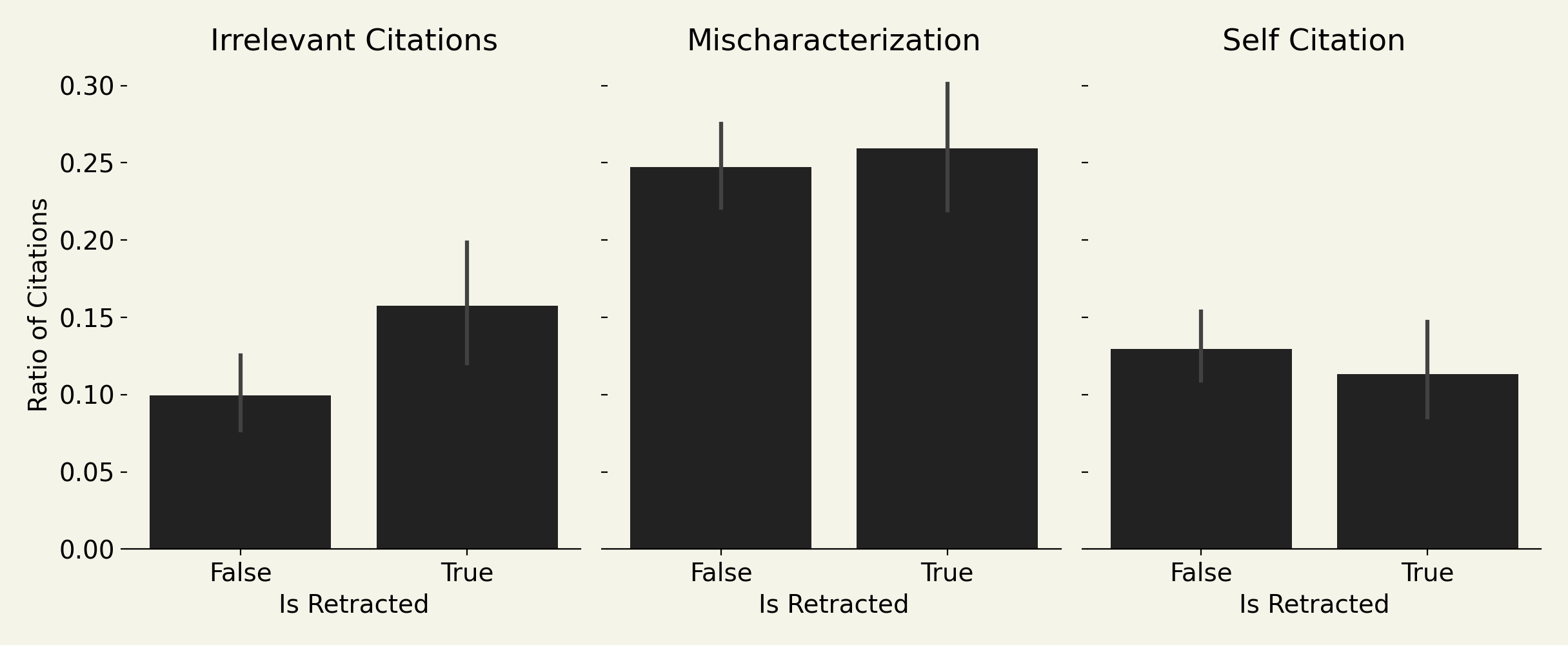
Now here are the results on using these as signals to predict retracted papers:
The p-values of the three signals:
- self-citations. p = 0.402
- citations to irrelevant papers. p = 0.016
- citations that are mischaracterizations of the cited work. p = 0.618
Self-citations are not predictive of an article being retracted. It turns out, people just reliably self-cite at about 10%. Retracted papers even self-cite a bit less than non-retracted papers.
Retracted papers do not mischaracterize cited articles any more than others. This is kind of interesting - it basically says that retracted articles are not any more sloppy or disingenuous when presenting other work. No signal here.
Retracted papers have more irrelevant citations. This is the best signal in the end. It is disappointing to me because there is no "mechanistic" connection to scientific fraud. Some of the papers listed in this retracted paper dataset include ones from image duplication, fake data, faked peer review, and citation rings. None of these seem tightly connected to irrelevant citations, except I guess citation rings. Anyway - this is at least a new correlating signal for spotting articles that will likely be retracted.
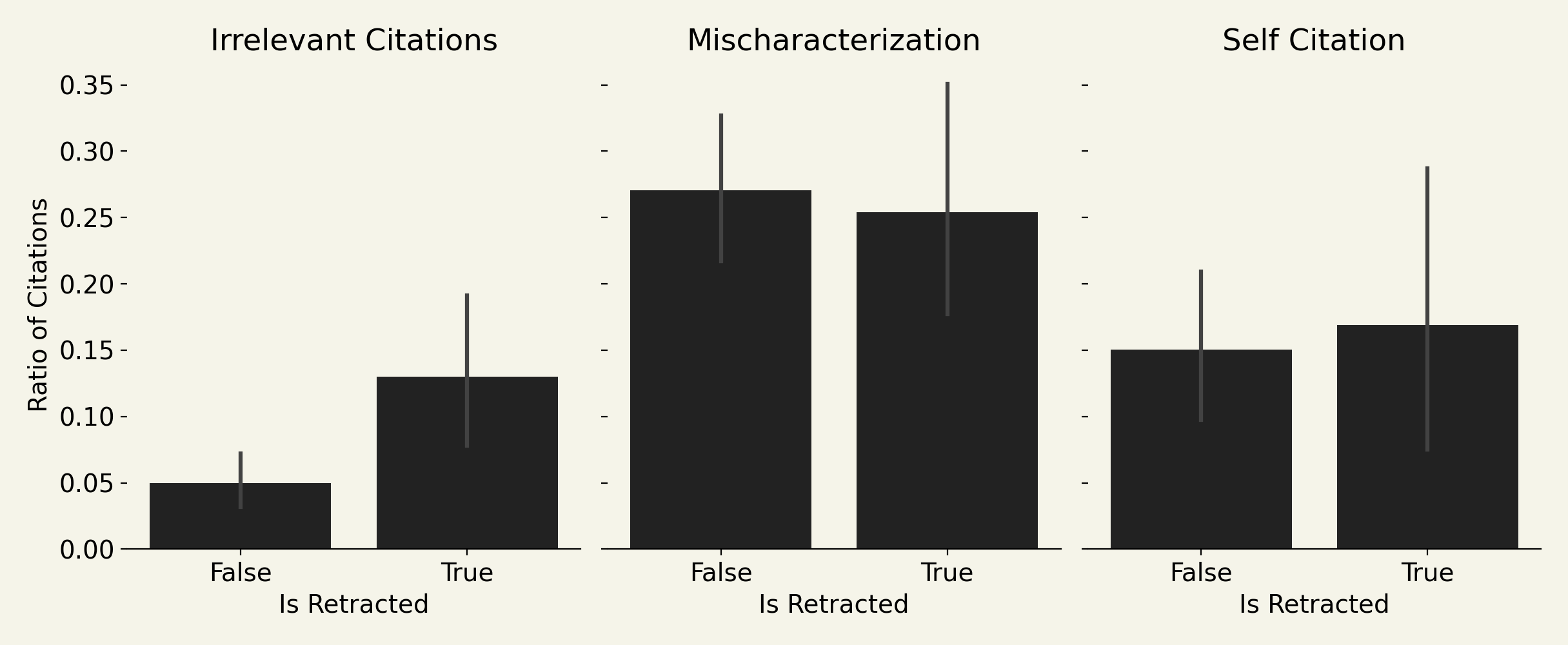
size effects
Since I'm only able to fetch ~20% of cited papers, we can repeat the analysis only on reference sets that we have at least 50% of the references checked. Like if an article has 100 references, we were able to get at least 50 downloaded to check against the claims in the paper. I wanted to check this bias because we had many papers with small fetched reference counts (e.g., ratios where the denominator is 2 or 3). This criteria yields only 84 articles (26 retracted) from the original 682.
The p-values of the three signals:
- self-citations. p = 0.306
- citations to irrelevant papers. p = 0.015
- citations that are mischaracterizations of the cited work. p = 0.762
The conclusions are thus unaffected by the sample size bias.
case study
Let's look at a famous example -- the retracted paper "High-Affinity Naloxone Binding to Filamin A Prevents Mu Opioid Receptor-Gs Coupling Underlying Opioid Tolerance and Dependence" by Hoau-Yan Wang. This paper was part of the basis for a clinical trial by an Alzheimer's research company called Cassava that ended up failing, likely because the scientific basis of the drug was based on retracted and fraudulent research.12 This paper was retracted on the basis of image duplication.
I was able to retrieve 23 of the 63 papers cited in the work. From these, I got:
- 19% for self-citations
- 4% for irrelevant citations
- 35% for mischaracterized citations
which means the signal of irrelevant citations would not flag this. Probably, we are picking up mostly citation rings or paper-mills in the signal, which is not a characteristic of this paper. Nevertheless, let's look at some of the mischaracterized citations.
First, the paper by Wang states:
"Ultra-low-dose opioid antagonists were initially thought to preferentially bind a subset of MORs [1], and a Gs-coupling MOR subpopulation was again recently proposed [9]."
These are about reference 1 ("Antagonists of excitatory opioid receptor functions enhance morphine's analgesic potency and attenuate opioid tolerance/dependence liability")13 and the model identifies these as mischaracterizations based on this reasoning:
Which is an interesting contrast - that it is not clearly μ opioid receptors (MOR) being bound by the antagonists.
Here's another one that shows a bit of weakness for this approach
and the model remarks that the reference mentioned only identifies 20, not over 30, and not including MOR. But, the model only has access to reference 14 here - so that including all references (13, 14, and 15) may have been required to understand the whole statement.
Here's another one
is considered a mischaracterization because
This one is more subtle - since MOR can couple to both Gi/Go and other Gs - so they may just be reporting on different conditions.
Anyway - the conclusion is that the method finds some interesting disagreements with cited papers, but would probably not flag this paper as fraudulent.
scaling to all literature
This method requires a long parsing step of an input paper and N calls over the cited papers with large language models. Assuming the N calls is the longest operation, we can try to compute the cost of implementing this in a few scenarios.
A typical research paper is 30,000 words and we need to ingest the entire paper (plus small excerpts from the paper we're checking against) The output from the language models is about 50 words (the choice and reasoning), so we can neglect that. If we take , we get a nice round number of about 1M words per research paper to check. Typical frontier models like deepseek's R1 and Gemini 1.5 pro 002 (used here) are around $1 per million input tokens. The number of articles in arXiv per month is bit under 25,000 and the total number of papers total published per month is about 500,000. If we wanted to check Wikipedia, a 2021 estimate put the number of external citations to scholarly sources at 4M.14 Thus, we get the following costs for each scenario:
| Scenario | Papers | Cost |
|---|---|---|
| arXiv submissions | 25,000 / mo | $25,000 / mo |
| published papers | 500,000 / mo | $500,000 / mo |
| all literature | 250M | $250M |
| wikipedia | 4M citations | $130,000 |
other people working on this
I recently found out about two other groups working on this:
-
yes no error - they have a pretty comprehensive pipeline to critique methods, figures, citations, etc. The interface looks cool and the whitepaper has a lot of details - would love to see some results though!
-
Black Spatula Proejct This one seems earlier, so not much to report, but is building open source tools for this.
I think this is an awesome topic and hope more progress gets made soon!
conclusions
Of the three possible signals for if a paper will be retracted, the only significant one was the number of cited claims that had irrelevant references. Like a cited claim - "biology is important[5]" - cites [5] but [5] is a paper about market theory. A typical paper has 5% of these. A retracted paper has >10%. Probably this is because the majority of retracted papers considered in my analysis arose from citation-rings/paper mills.
I ran this system over my own papers and it really did a good job of finding mistakes in citations. I had some published papers where both there was a mistake in citing the right paper, and I had misinterpreted a paper. A check like this could be really helpful, especially since it is pretty robust to document format/type.
I think it could also be interesting for funders, either governments or philanthropies. Not really as a check on correctness of a proposal - that requires experts - but it can give a signal of if a proposal has put some care into correctly characterizing the state of the field.
The scale is fascinating to consider. We can be running larger language models on all incoming papers and the articles they cite at a cost of $1 per paper. For reference, journals charge from about $1,000 to $13,000 to publish an article. So this would be a relatively small additional cost.
Why blog this instead of write a paper? Writing research papers is hard and these results are not very strong. I also want to put this out there to get more people thinking about these ideas. We may be able to make much stronger signals and catch fraud more easily soon with advances in AI.
code
This relies a lot on PaperQA, whose source is released on github: github.com/future-House/paper-qa. But the PaperQA API/DB which simplifies fetching open access/preprinted full-text papers is not available because (1) only like 30% of papers can be found from pre-prints/open access links and it is really bad in specific fields like economics or politics that do not use pre-prints and (2) we don't have a platform yet that can handle rate limits from external users.
Footnotes
-
There is no plausible reason for why you would duplicate pieces of images and there are few deliberations. If there are clear duplicates, there was fraud. Nevertheless, it can take years and involve lawsuits for researchers to be held accountable and publications be retracted. If duplicating images wasn't such a low false positive method, I'm afraid we would in an even worse state than today with scientific fraud detection. ↩
-
Example https://datacolada.org/98 ↩
-
Richardson, Reese A. K., et al. "Widespread Misidentification of SEM Instruments in the Peer-Reviewed Materials Science and Engineering Literature." OSF Preprints, 27 Aug. 2024, https://doi.org/10.31219/osf.io/4wqcr. ↩
-
It was 28%, but you can also take 22% as the number if you account for papers that had multiple microscopes and plausibly could have mixed them. I took 25%, which is in between these. ↩
-
Bik, Elisabeth M., et al. "Analysis and correction of inappropriate image duplication: the molecular and cellular biology experience." Molecular and cellular biology (2018). https://doi.org/10.1128/MCB.00309-18 ↩ ↩2
-
For example, as assessed by Finnish Publication Forum: https://julkaisufoorumi.fi/en ↩
-
Begley, C. Glenn, and Lee M. Ellis. "Raise standards for preclinical cancer research." Nature 483.7391 (2012): 531-533. https://www.nature.com/articles/483531a; Freedman, Leonard P., Iain M. Cockburn, and Timothy S. Simcoe. "The economics of reproducibility in preclinical research." PLoS biology 13.6 (2015): e1002165. https://doi.org/10.1371/journal.pbio.1002165, ↩
-
David Bimler. Better Living through Coordination Chemistry: A descriptive study of a prolific papermill that combines crystallography and medicine, 15 April 2022, PREPRINT (Version 1) available at Research Square. https://www.researchsquare.com/article/rs-1537438/v1 ↩
-
RetractionWatch retracted article DB: https://gitlab.com/crossref/retraction-watch-data/-/blob/main/retraction_watch.csv?ref_type=heads ↩
-
gemini-pro-002 on vertex-ai ↩
-
Piller, Charles. "Co-developer of Cassava’s Potential Alzheimer’s Drug Cited for ‘Egregious Misconduct’." ScienceInsider, 12 Oct. 2023, https://www.science.org/content/article/co-developer-cassava-s-potential-alzheimer-s-drug-cited-egregious-misconduct ↩
-
Crain, Stanley M., and Ke-Fei Shen. "Antagonists of excitatory opioid receptor functions enhance morphine's analgesic potency and attenuate opioid tolerance/dependence liability." PAIN® 84.2-3 (2000): 121-131. https://doi.org/10.1016/S0304-3959(99)00223-7 ↩
-
Singh, Harshdeep, Robert West, and Giovanni Colavizza. "Wikipedia citations: A comprehensive data set of citations with identifiers extracted from English Wikipedia." Quantitative Science Studies 2.1 (2021): 1-19. https://doi.org/10.1162/qss_a_00105 ↩